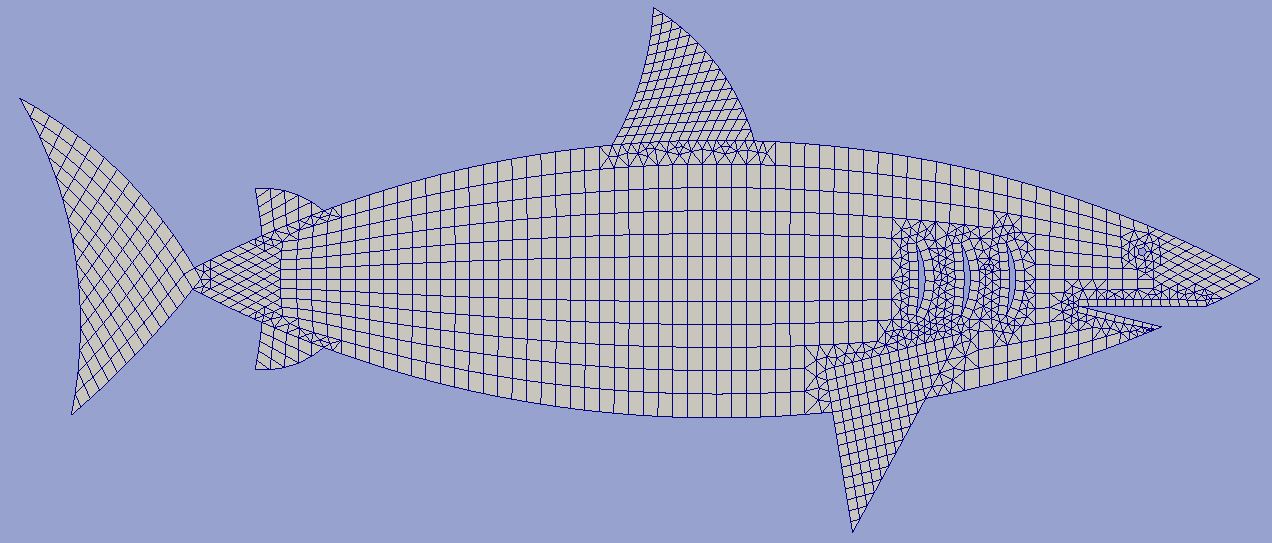
Example 4¶
Using mappings for grid construction
from hybmeshpack import hmscript as hm
hm.check_compatibility("0.5.0", 2)
ncirc = 256 # precision of circle contours
# I. Build body parts grids ***************************************************
# build body contour as intersection of two circles
circ1 = hm.add_circ_contour3([6, 0], [-6, 0], 0.08, ncirc)
circ2 = hm.add_circ_contour3([-6, 0], [6, 0], 0.08, ncirc)
bodyc = hm.clip_domain(circ1, circ2, "intersection")
# assemble a prototype grid for body by attaching triangles to left and
# right side of a rectangle.
ntri, nspan = 12, 60 # vertical and horizontal partition of rectangle
g3l = hm.add_triangle_grid([0, 0], [-0.5, 0.5], [0, 1], ntri)
g3r = hm.add_triangle_grid([5, 0], [5.5, 0.5], [5, 1], ntri)
g4 = hm.add_unf_rect_grid([0, 0], [5, 1], nspan, ntri)
# Grids in triangle and rectangle areas have same partition at contact line
# so we can simply unite them with zero buffer size
gb = hm.unite_grids(g4, [(g3l, 0), (g3r, 0)])
# Map grid at hexagon area on body area so that points at the acute angles
# of base grid contour be translated into acute angle vertices of body contour.
# We use snap="no" since there is no need to preserve initial body contour
# precisely.
body = hm.map_grid(gb, bodyc, [[-0.5, 0.5], [5.5, 0.5]],
[[-6, 0], [6, 0]], snap="no")
# build tail area by clipping of three circle areas
circ1 = hm.add_circ_contour3([0, 0], [-2, 2], 0.2, ncirc)
circ2 = hm.add_circ_contour3([-2.5, -2.3], [0, 0], 0.1, ncirc)
circ3 = hm.add_circ_contour3([-1.5, -1.8], [-2, 2], 0.2, ncirc)
d1 = hm.clip_domain(circ1, circ2, "intersection")
tailc = hm.clip_domain(d1, circ3, "difference")
# using triangle as a prototype grid for mapping
g3 = hm.add_triangle_grid([-1, 0], [0, 2], [1, 0], 15)
tail = hm.map_grid(
g3, tailc,
[[-1, 0], [0, 2], [1, 0]],
[[-2, 2], [0, 0], [-2.5, -2.3]], snap="no")
hm.move_geom([tail], -5.7, 0)
# upper fin area is a assembled from two circles and meshed body area
circ1 = hm.add_circ_contour3([0.5, 1], [-0.7, 3.0], 0.4, ncirc)
circ2 = hm.add_circ_contour3([-1.5, 1], [-0.7, 3.0], 0.3, ncirc)
d1 = hm.clip_domain(circ1, circ2, "difference")
fin1c = hm.clip_domain(d1, body, "difference")
# Mapping using the same triangle as we've used for tail mapping.
# we give only approximate values: [-1.5, 1.3], [0.5, 1.3].
# To ensure that given coordinates will be project to angular vertices
# we use project_to="corner" option
fin1 = hm.map_grid(
g3, fin1c,
[[1, 0], [0, 2], [-1, 0]],
[[-0.7, 3], [-1.5, 1.3], [0.5, 1.3]], snap="no", project_to="corner")
# side fin is a simple triangle
fin2 = hm.add_triangle_grid([2.7, -0.6], [1.2, -1.0], [1.5, -2.8], 15)
# back fins assemble is similar to upper fin assemble
circ1 = hm.add_circ_contour2([-5.1, -1], [-5.0, 0], [-5.1, 1], ncirc)
circ2 = hm.add_circ_contour2([-5.1, -1], [-4, 0], [-5.1, 1], ncirc)
d1 = hm.clip_domain(circ2, circ1, "difference")
fin34c = hm.clip_domain(d1, body, "difference")
g3 = hm.add_triangle_grid([1, 0], [0, 0], [0, 1], 5)
fin3 = hm.map_grid(
g3, fin34c,
[[1, 0], [0, 0], [0, 1]],
[[-4, 0.5], [-5.1, 0.3], [-5.1, 1]], project_to="corner", snap="no")
fin4 = hm.map_grid(
g3, fin34c,
[[0, 0], [1, 0], [0, 1]],
[[-5.2, -0.3], [-5.1, -1], [-4.1, -0.7]], project_to="corner", snap="no")
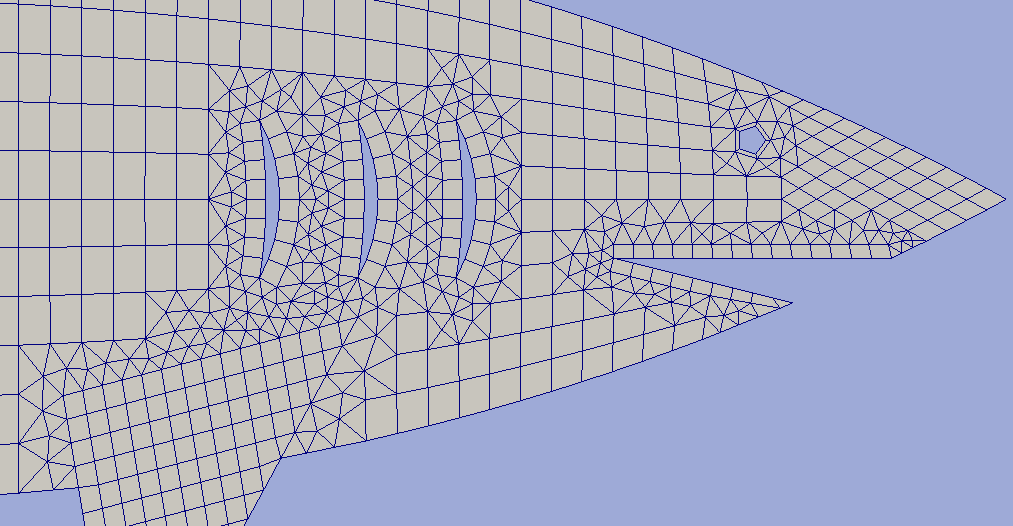
# jaw is constructed by excluding triangle from the body grid area and
# constructing a boundary grid to get rid of badly shaped cells near the cut
jcont = hm.create_contour([[4, -0.3], [8, -1.3], [8, -0.3], [4, -0.3]])
body = hm.exclude_contours(body, jcont, "inner")
btopt = hm.BoundaryGridOptions(
body, [0, 0.07], "left", 0.1,
start_point=[5, -0.5], end_point=[5.45, -0.36], project_to="corner")
# shut down acute angle algorithm in order to obtain grid which lies strictly
# within body contour at the bottom point
btopt.range_angles[0] = 0
jgrid = hm.build_boundary_grid(btopt)
# gills areas are constructed by a boundary grid built around
# contours which are constructed by two circles intersection
circ1 = hm.add_circ_contour3([2.7, -0.4], [2.7, 0.4], 0.4, ncirc)
circ2 = hm.add_circ_contour3([2.7, -0.4], [2.7, 0.4], 1.2, ncirc)
gillc = hm.clip_domain(circ2, circ1, "difference")
gillc = hm.partition_contour(gillc, "const", 0.1)
gills = hm.build_boundary_grid1(gillc, [0, 0.1], "right")
[gills2, gills3] = hm.copy_geom([gills] * 2)
hm.move_geom(gills2, -0.5, 0)
hm.move_geom(gills3, 0.5, 0)
# since gills grids have no intersection area, their union
# does nothing but assembling three grids into single connectivity table.
gills = hm.unite_grids(gills, [(gills2, 0), (gills3, 0)])
# an eye is built as a ring grid.
eye = hm.add_unf_ring_grid([4.7, 0.3], 0.05, 0.1, 5, 1)
# II. Coupling grids **********************************************************
# Fins and the body are not tightly connected and direct superposition
# procedure may give bad results. To fix that we snap body grid to those fins.
p1 = hm.get_point(fin1, vclosest=[-1.2, 1.4])
p2 = hm.get_point(fin1, vclosest=[0.5, 1.4])
body = hm.snap_grid_to_contour(body, fin1, p2, p1, p1, p2)
p1 = hm.get_point(fin3, vclosest=[-5.1, 0.4])
p2 = hm.get_point(fin3, vclosest=[-4.3, 0.7])
body = hm.snap_grid_to_contour(body, fin3, p2, p1, p1, p2)
p1 = hm.get_point(fin4, vclosest=[-5.1, -0.4])
p2 = hm.get_point(fin4, vclosest=[-4.3, -0.7])
body = hm.snap_grid_to_contour(body, fin4, p1, p2, p2, p1)
# Now fin and body grid domains have intersection and can be united.
# Use zero_angle_approx=10 to let buffer reset boundary points
shark = hm.unite_grids(body, [(fin1, 0.1), (fin3, 0.1), (fin4, 0.1)],
zero_angle_approx=10)
# all other grids have clear intersections and could be united as a chain.
# we use empty_holes=True in order to preserve hulls at eye and gills areas.
shark = hm.unite_grids(
shark, [(gills, 0.1), (tail, 0.1), (eye, 0.1), (jgrid, 0.1), (fin2, 0.15)],
empty_holes=True, zero_angle_approx=10)
# leave only resulting grid
hm.remove_all_but(shark)
# boundary grids around gills contain hanging boundary nodes. To get rid of
# them we use heal_grid procedure with default parameters
hm.heal_grid(shark)
# check grid skewness
if not hm.skewness(shark)['ok']:
print "Grid contains bad cells"
# exporting to vtk
hm.export_grid_vtk(shark, "shark.vtk")